Chapter 3 Data Manipulation in R
This chapter will take you through a brief walk through of cleaning some data in R. When starting out with R, it’s not important that you memorize every command and learn every line, but rather that you start to think about how a computer programmer would solve a question (you!) rather than get an undergrad RA to fix it by hand.
The first thing that you want to do to any script is to put a header on it. It should cover a few things such as
- A Title
- Who wrote the script?
- When did you edit it last?
- What does it do?
- How does it work?
- Does it take anything in specific?
The more information you can provide, the better. Remember that 6-Month-From-You now is going to be either your best or worst collaborator.
Each script in R out to follow a general pattern. First list out some information about it, then load in your libraries, and then set your working directory and load your data.
## Warning: package 'stringr' was built under R version 3.4.3## Warning: package 'data.table' was built under R version 3.4.4Next thing you would do is set your working directory and load in your data. Note here that you need to tell R exactly where to look!
# Make sure to load in both datasets as we will want them both in our analysis.
# We also want to make sure to clean both datasets as we are going.
experiment.data <- read.csv("datasets/Demographic_Data.csv")
item.level.data <- read.csv("datasets/ItemLevelData.csv")You can also import data using RStudio’s ‘Import Data’ function in the top right corner.
Import Data via RStudio’s GUI
If you do do that, make sure to add in any of the libraries and code in your script that is printed out below in RStudio’s consle!
Next you should familiarize yourself with what your data looks like. Not only do you have to be cautious of the shape of your data (wide vs. long), but there are a few different types of data that R uses in its functionality.
Looking at the dataset from this experiment, we see lots of different types of data listed. R did its best to guess what ‘kind’ of data each one is, but sometimes you have to change data types.
## 'data.frame': 251 obs. of 67 variables:
## $ Sub : int 10 11 12 13 14 15 16 17 18 19 ...
## $ ACTIVE : int 38 37 39 33 48 47 55 42 36 36 ...
## $ PERCEPTUAL : int 47 47 49 42 57 55 60 61 43 36 ...
## $ MUSICAL : int 28 16 21 26 7 8 46 39 19 24 ...
## $ SINGING : int 33 22 40 19 24 24 45 25 29 28 ...
## $ EMOTIONS : int 39 32 30 34 35 36 42 41 33 24 ...
## $ GENERAL : int 75 67 82 68 60 65 118 86 69 68 ...
## $ inst : Factor w/ 70 levels "","air drums",..: 9 4 67 46 28 36 36 36 48 28 ...
## $ Age : int 29 22 19 23 23 19 19 21 19 21 ...
## $ Gender : Factor w/ 13 levels "","COUNTRY","f",..: 6 6 6 12 12 4 6 4 12 10 ...
## $ Major : Factor w/ 147 levels "","AGRIBUSINESS",..: 24 126 38 88 67 79 95 17 143 47 ...
## $ Minor : Factor w/ 65 levels ""," Economics",..: 29 NA 50 NA 4 NA NA NA NA 39 ...
## $ SchoolUGorG : int 1 1 1 1 1 1 1 1 1 1 ...
## $ MuscMajMinor : int 1 1 1 1 1 1 2 1 1 1 ...
## $ BeginTrain : Factor w/ 20 levels "","10","11","12",..: 20 2 2 3 1 1 19 16 4 16 ...
## $ AbsPitch : Factor w/ 7 levels "","no","No","NO",..: 4 2 4 4 4 4 2 4 4 2 ...
## $ YearsPrac : int 4 1 3 7 0 0 11 7 2 6 ...
## $ PeakPrac : int 5 2 1 1 0 0 3 2 2 1 ...
## $ Live12Mo : Factor w/ 23 levels "","0","1","10",..: 9 15 3 2 20 9 17 12 14 18 ...
## $ TheoryTrain : int 4 0 0 0 0 0 4 4 0 0 ...
## $ FormalYrs : int 4 1 3 7 0 0 11 8 2 2 ...
## $ NoOfInst : int 1 0 1 2 0 0 5 1 1 0 ...
## $ ActListenMin : Factor w/ 34 levels "","0","1","10",..: 30 30 9 24 5 30 9 24 24 22 ...
## $ SubjectNo : int 10 11 12 13 14 15 16 17 18 19 ...
## $ SubRaven : int 10 11 12 13 14 15 16 17 18 19 ...
## $ RavenB1 : int 10 7 3 10 7 6 8 8 4 3 ...
## $ RavenB2 : int 8 10 7 10 8 10 9 10 6 3 ...
## $ RavenB3 : int 10 6 7 11 5 6 8 8 5 3 ...
## $ RavenTotal : int 28 23 17 31 20 22 25 26 15 9 ...
## $ OspanSub : int 10 11 12 13 14 15 16 17 18 19 ...
## $ OspanAccError : int 8 7 7 1 2 12 4 8 5 37 ...
## $ OspanMathError : int 12 12 7 2 2 15 4 8 8 37 ...
## $ OspanAbsoluteScore: int 29 20 48 68 33 20 68 36 16 14 ...
## $ OspanPartialScore : int 47 41 65 73 50 44 70 64 30 23 ...
## $ OspanB1 : int 21 11 21 25 17 14 25 19 10 8 ...
## $ OspanB2 : int 14 18 23 23 16 17 20 24 11 6 ...
## $ OspanB3 : int 12 12 21 25 17 13 25 21 9 9 ...
## $ OspanSpeedErrTot : int 4 5 0 1 0 3 0 0 3 0 ...
## $ SymSub : int 10 11 12 13 14 15 16 17 18 19 ...
## $ SymAccErrTot : int 1 4 2 1 0 4 2 3 2 23 ...
## $ SymSpeedErrTot : int 0 1 0 0 0 1 0 0 0 0 ...
## $ SspanAbsScore : int 10 15 21 27 15 9 30 24 12 18 ...
## $ SspanPartiaScore : int 19 29 34 34 26 25 38 30 18 24 ...
## $ SspanB1 : int 8 11 10 13 8 9 13 7 8 8 ...
## $ SspanB2 : int 4 13 13 10 8 10 14 14 4 7 ...
## $ SspanB3 : int 7 5 11 11 10 6 11 9 6 9 ...
## $ SymmErrTot : int 1 5 2 1 0 5 2 3 2 23 ...
## $ ToneSub : int 10 11 12 13 14 15 16 17 18 19 ...
## $ ToneAccErrTot : int 8 5 8 1 4 7 3 6 3 36 ...
## $ ToneMathErrTot : int 9 5 8 2 5 10 5 6 6 38 ...
## $ TspanAbsScore : int 9 21 27 37 10 4 23 14 3 0 ...
## $ TspanPartScore : int 36 47 55 63 31 34 58 48 37 26 ...
## $ TspanB1 : int 13 14 17 20 8 9 20 12 9 9 ...
## $ TspanB2 : int 6 19 21 22 9 12 19 18 12 5 ...
## $ TspanB3 : int 17 14 17 21 14 13 19 18 16 12 ...
## $ ToneSpeedErrTot : int 1 0 0 1 1 3 2 0 3 2 ...
## $ BeatSub : int 10 11 12 13 14 15 16 17 18 19 ...
## $ BeatTotal : int 11 10 14 13 14 10 14 11 7 12 ...
## $ BeatAcc : num 0.61 0.56 0.78 0.72 0.78 0.56 0.78 0.61 0.39 0.67 ...
## $ MelodicSub : int 10 11 12 13 14 15 16 17 18 19 ...
## $ MelodicTot : int 7 7 6 10 5 5 6 7 7 7 ...
## $ MelodicAcc : num 0.54 0.54 0.46 0.77 0.38 0.38 0.46 0.54 0.54 0.54 ...
## $ Native : int 1 1 1 1 1 1 1 2 1 1 ...
## $ Paid : int 1 1 1 1 1 1 1 1 1 1 ...
## $ jv_NoGMSI : int 0 0 0 0 0 0 0 0 0 0 ...
## $ jv_nonNative : int 0 0 0 0 0 0 0 1 0 0 ...
## $ jv_noOspan : int 0 0 0 0 0 0 0 0 0 0 ...Using read.csv() with its default setting, R had a couple of bad guesses on variables we might need.
We will have to reassign the variable types or else we’ll run into trouble later.
In R we use factor for grouping and analysis, best practice is to not set it as that
until you are OK with the format.
It’s easist to manipulate a character or string.
There are much more elegant ways to do this, but just to make a point, I’ve gone through and fixed each one manually.
Notice how the output of str() has changed.
experiment.data$inst <- as.character(experiment.data$inst)
experiment.data$Gender <- as.character(experiment.data$Gender)
experiment.data$Major <- as.character(experiment.data$Major)
experiment.data$Minor <- as.character(experiment.data$Minor)
experiment.data$BeginTrain <- as.character(experiment.data$BeginTrain)
experiment.data$AbsPitch <- as.character(experiment.data$AbsPitch)
experiment.data$Live12Mo <- as.numeric(experiment.data$Live12Mo)
experiment.data$ActListenMin <- as.character(experiment.data$ActListenMin)
str(experiment.data) # Notice how that our character columns now have " " around them.## 'data.frame': 251 obs. of 67 variables:
## $ Sub : int 10 11 12 13 14 15 16 17 18 19 ...
## $ ACTIVE : int 38 37 39 33 48 47 55 42 36 36 ...
## $ PERCEPTUAL : int 47 47 49 42 57 55 60 61 43 36 ...
## $ MUSICAL : int 28 16 21 26 7 8 46 39 19 24 ...
## $ SINGING : int 33 22 40 19 24 24 45 25 29 28 ...
## $ EMOTIONS : int 39 32 30 34 35 36 42 41 33 24 ...
## $ GENERAL : int 75 67 82 68 60 65 118 86 69 68 ...
## $ inst : chr "clarinet" "bass" "Voice" "saxophone" ...
## $ Age : int 29 22 19 23 23 19 19 21 19 21 ...
## $ Gender : chr "FEMALE" "FEMALE" "FEMALE" "MALE" ...
## $ Major : chr "BUSINESS MANAGEMENT" "psychology" "COMMUNICATION STUDIES" "Mechanical Engineering" ...
## $ Minor : chr "HUMAN RESOURCE MANAGEMENT" NA "SOCIAL WORK" NA ...
## $ SchoolUGorG : int 1 1 1 1 1 1 1 1 1 1 ...
## $ MuscMajMinor : int 1 1 1 1 1 1 2 1 1 1 ...
## $ BeginTrain : chr "9" "10" "10" "11" ...
## $ AbsPitch : chr "NO" "no" "NO" "NO" ...
## $ YearsPrac : int 4 1 3 7 0 0 11 7 2 6 ...
## $ PeakPrac : int 5 2 1 1 0 0 3 2 2 1 ...
## $ Live12Mo : num 9 15 3 2 20 9 17 12 14 18 ...
## $ TheoryTrain : int 4 0 0 0 0 0 4 4 0 0 ...
## $ FormalYrs : int 4 1 3 7 0 0 11 8 2 2 ...
## $ NoOfInst : int 1 0 1 2 0 0 5 1 1 0 ...
## $ ActListenMin : chr "60" "60" "180" "45" ...
## $ SubjectNo : int 10 11 12 13 14 15 16 17 18 19 ...
## $ SubRaven : int 10 11 12 13 14 15 16 17 18 19 ...
## $ RavenB1 : int 10 7 3 10 7 6 8 8 4 3 ...
## $ RavenB2 : int 8 10 7 10 8 10 9 10 6 3 ...
## $ RavenB3 : int 10 6 7 11 5 6 8 8 5 3 ...
## $ RavenTotal : int 28 23 17 31 20 22 25 26 15 9 ...
## $ OspanSub : int 10 11 12 13 14 15 16 17 18 19 ...
## $ OspanAccError : int 8 7 7 1 2 12 4 8 5 37 ...
## $ OspanMathError : int 12 12 7 2 2 15 4 8 8 37 ...
## $ OspanAbsoluteScore: int 29 20 48 68 33 20 68 36 16 14 ...
## $ OspanPartialScore : int 47 41 65 73 50 44 70 64 30 23 ...
## $ OspanB1 : int 21 11 21 25 17 14 25 19 10 8 ...
## $ OspanB2 : int 14 18 23 23 16 17 20 24 11 6 ...
## $ OspanB3 : int 12 12 21 25 17 13 25 21 9 9 ...
## $ OspanSpeedErrTot : int 4 5 0 1 0 3 0 0 3 0 ...
## $ SymSub : int 10 11 12 13 14 15 16 17 18 19 ...
## $ SymAccErrTot : int 1 4 2 1 0 4 2 3 2 23 ...
## $ SymSpeedErrTot : int 0 1 0 0 0 1 0 0 0 0 ...
## $ SspanAbsScore : int 10 15 21 27 15 9 30 24 12 18 ...
## $ SspanPartiaScore : int 19 29 34 34 26 25 38 30 18 24 ...
## $ SspanB1 : int 8 11 10 13 8 9 13 7 8 8 ...
## $ SspanB2 : int 4 13 13 10 8 10 14 14 4 7 ...
## $ SspanB3 : int 7 5 11 11 10 6 11 9 6 9 ...
## $ SymmErrTot : int 1 5 2 1 0 5 2 3 2 23 ...
## $ ToneSub : int 10 11 12 13 14 15 16 17 18 19 ...
## $ ToneAccErrTot : int 8 5 8 1 4 7 3 6 3 36 ...
## $ ToneMathErrTot : int 9 5 8 2 5 10 5 6 6 38 ...
## $ TspanAbsScore : int 9 21 27 37 10 4 23 14 3 0 ...
## $ TspanPartScore : int 36 47 55 63 31 34 58 48 37 26 ...
## $ TspanB1 : int 13 14 17 20 8 9 20 12 9 9 ...
## $ TspanB2 : int 6 19 21 22 9 12 19 18 12 5 ...
## $ TspanB3 : int 17 14 17 21 14 13 19 18 16 12 ...
## $ ToneSpeedErrTot : int 1 0 0 1 1 3 2 0 3 2 ...
## $ BeatSub : int 10 11 12 13 14 15 16 17 18 19 ...
## $ BeatTotal : int 11 10 14 13 14 10 14 11 7 12 ...
## $ BeatAcc : num 0.61 0.56 0.78 0.72 0.78 0.56 0.78 0.61 0.39 0.67 ...
## $ MelodicSub : int 10 11 12 13 14 15 16 17 18 19 ...
## $ MelodicTot : int 7 7 6 10 5 5 6 7 7 7 ...
## $ MelodicAcc : num 0.54 0.54 0.46 0.77 0.38 0.38 0.46 0.54 0.54 0.54 ...
## $ Native : int 1 1 1 1 1 1 1 2 1 1 ...
## $ Paid : int 1 1 1 1 1 1 1 1 1 1 ...
## $ jv_NoGMSI : int 0 0 0 0 0 0 0 0 0 0 ...
## $ jv_nonNative : int 0 0 0 0 0 0 0 1 0 0 ...
## $ jv_noOspan : int 0 0 0 0 0 0 0 0 0 0 ...The next thing we’d want to check in our data cleaning is for any sort of import errors.
We can use combinations of the functions names(),table(), is.na(), and complete.cases() to get a brief summary of what is missing.
You could also use R’s plotting funcitonality to see if there are any participants with weird subject numbers or maybe negative values where there should not be.
If you find any problems, code in the solution in R!
Resist the urge to fix it in the Excel file!
# Check for Import Errors
table(complete.cases(experiment.data)) # Not all observations have everything! ##
## FALSE TRUE
## 74 177## [1] TRUE FALSE TRUE FALSE TRUE FALSE FALSE FALSE FALSE TRUE FALSE
## [12] FALSE FALSE FALSE TRUE FALSE TRUE FALSE FALSE TRUE TRUE FALSE
## [23] TRUE FALSE FALSE TRUE FALSE FALSE TRUE FALSE FALSE TRUE TRUE
## [34] FALSE TRUE FALSE TRUE FALSE TRUE FALSE FALSE FALSE TRUE TRUE
## [45] TRUE FALSE FALSE TRUE TRUE TRUE FALSE FALSE TRUE FALSE FALSE
## [56] FALSE TRUE FALSE TRUE FALSE FALSE TRUE FALSE TRUE FALSE FALSE
## [67] FALSE TRUE FALSE TRUE FALSE FALSE FALSE FALSE FALSE TRUE TRUE
## [78] FALSE FALSE FALSE TRUE TRUE TRUE TRUE FALSE TRUE TRUE FALSE
## [89] FALSE FALSE FALSE TRUE FALSE FALSE FALSE TRUE FALSE TRUE TRUE
## [100] TRUE FALSE FALSE FALSE FALSE FALSE FALSE TRUE FALSE FALSE FALSE
## [111] TRUE FALSE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE
## [122] TRUE FALSE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE
## [133] TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE
## [144] TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE
## [155] TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE
## [166] TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE
## [177] TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE FALSE TRUE TRUE
## [188] TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE
## [199] TRUE FALSE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE
## [210] TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE
## [221] TRUE TRUE TRUE TRUE TRUE FALSE FALSE TRUE FALSE TRUE TRUE
## [232] TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE
## [243] TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE##
## FALSE TRUE
## 16723 94Later we’ll cover how to change individual cases, but as I will note, you will want to avoid hard coding in any fixes if you can. Let’s first look at how we might fix a messy gender problem.
3.1 Cleaning Response Data
In your data cleaning before you might have noticed that participants were able to freely respond with whatever gender they wanted. Most data look to fall within the normal binary, but the computer needs things to be exactly the same before making an easy split? What would be the laziest, most effecient way to fix the gender column? What format does the variable have to be in order to make the changes that you need? When you have it figured out, make sure to run the code from top to bottom to make sure things go in the right order!!! As we are not dealing with huge amounts of data, the table() function will help out.
Let’s now take a look at some of these problem ones Why, for example is Begin Training not working? Print the variable to see.
## [1] "9" "10" "10" "11" ""
## [6] "" "8" "6" "12" "6"
## [11] "" "" "10" "" "7"
## [16] "16" "10" "18" "15" "7"
## [21] "8" "10" "11" "8" "12"
## [26] "6" "" "17" "" ""
## [31] "10" "15" "15" "8" "6"
## [36] "13" "16" "" "7" "7"
## [41] "10" "" "12" "5" "11"
## [46] "9" "" "21" "" "16"
## [51] "17" "11" "" "13" ""
## [56] "15" "6" "15" "9" "16"
## [61] "10" "18" "" "11" "9"
## [66] "12" "10" "4" "" "9"
## [71] "9" "" "14" "11" "10"
## [76] "7" "" "17" "9" "16"
## [81] "7" "7" "5" "" ""
## [86] "10" "10" "9" "14" "17"
## [91] "11" "" "" "12" "6"
## [96] "14" "8" "11" "" ""
## [101] "12" "6" "10" "14" "8"
## [106] "13" "12" "8" "11" "8"
## [111] "14" "11" "8" "" ""
## [116] "10" "13" "13" "5" "15"
## [121] "" "11" "8" "12" "10"
## [126] "9" "6" "7" "10" "6"
## [131] "8" "" "16" "" ""
## [136] "" "11" "12" "7" ""
## [141] "4" "7" "7" "" "10"
## [146] "10" "12" "10" "" "12"
## [151] "12" "6" "14" "7" ""
## [156] "6" "18" "8" "12" "15"
## [161] "13" "" "10" "11" "10"
## [166] "13" "8" "" "6" "8"
## [171] "8" "" "9" "13" ""
## [176] "16" "10" "9" "10" "5"
## [181] "11" "" "15" "11" "13"
## [186] "14" "11" "2" "12" "11"
## [191] "8" "13" "8" "9" "12"
## [196] "9" "7" "12" "9" "8"
## [201] "9" "6" "11" "11" "9"
## [206] "10" "12" "12" "10" "6"
## [211] "9" "14" "5" "11" "12"
## [216] "8" "9" "11" "5" "13"
## [221] "10" "11" "9" "9" "10"
## [226] "5" "8" "10" "7" "10"
## [231] "12" "10" "7" "13" "11"
## [236] "11" "10" "9" "8" "12"
## [241] "11" "7" "10" "12" "29"
## [246] "5" "13" "9" "9" "7th grade"
## [251] "8"Some people didn’t respond, one person decided to tell us what grade they started. There are 2 ways to go about fixing this. We could “hardcode” the problem if this is the only time we will do this analysis on this dataset or we could try to write a line of code that doesn’t care what exact position the error is. On line 250 in this object is the thing that needs swapping out. We can access it with R’s indexing. Counting from index we see it’s in line 250.
## [1] "7th grade"Quick, ask yourself why we don’t use the comma here?! If you were set on using the comma, what would you change? Ok, now let’s swap in the value we want with <- Remember we are putting a value into a character operator so it has to have ""
## [1] "9" "10" "10" "11" "" "" "8" "6" "12" "6" "" "" "10" ""
## [15] "7" "16" "10" "18" "15" "7" "8" "10" "11" "8" "12" "6" "" "17"
## [29] "" "" "10" "15" "15" "8" "6" "13" "16" "" "7" "7" "10" ""
## [43] "12" "5" "11" "9" "" "21" "" "16" "17" "11" "" "13" "" "15"
## [57] "6" "15" "9" "16" "10" "18" "" "11" "9" "12" "10" "4" "" "9"
## [71] "9" "" "14" "11" "10" "7" "" "17" "9" "16" "7" "7" "5" ""
## [85] "" "10" "10" "9" "14" "17" "11" "" "" "12" "6" "14" "8" "11"
## [99] "" "" "12" "6" "10" "14" "8" "13" "12" "8" "11" "8" "14" "11"
## [113] "8" "" "" "10" "13" "13" "5" "15" "" "11" "8" "12" "10" "9"
## [127] "6" "7" "10" "6" "8" "" "16" "" "" "" "11" "12" "7" ""
## [141] "4" "7" "7" "" "10" "10" "12" "10" "" "12" "12" "6" "14" "7"
## [155] "" "6" "18" "8" "12" "15" "13" "" "10" "11" "10" "13" "8" ""
## [169] "6" "8" "8" "" "9" "13" "" "16" "10" "9" "10" "5" "11" ""
## [183] "15" "11" "13" "14" "11" "2" "12" "11" "8" "13" "8" "9" "12" "9"
## [197] "7" "12" "9" "8" "9" "6" "11" "11" "9" "10" "12" "12" "10" "6"
## [211] "9" "14" "5" "11" "12" "8" "9" "11" "5" "13" "10" "11" "9" "9"
## [225] "10" "5" "8" "10" "7" "10" "12" "10" "7" "13" "11" "11" "10" "9"
## [239] "8" "12" "11" "7" "10" "12" "29" "5" "13" "9" "9" "12" "8"Nice, no more text data, but what if it’s not always in 250? For example, what do we do with all these blank spaces? Let’s use R’s inbuilt ifelse() function to go through this vector and swap out what we want!
## [1] "9" "10" "10" "11" "0" "0" "8" "6" "12" "6" "0" "0" "10" "0"
## [15] "7" "16" "10" "18" "15" "7" "8" "10" "11" "8" "12" "6" "0" "17"
## [29] "0" "0" "10" "15" "15" "8" "6" "13" "16" "0" "7" "7" "10" "0"
## [43] "12" "5" "11" "9" "0" "21" "0" "16" "17" "11" "0" "13" "0" "15"
## [57] "6" "15" "9" "16" "10" "18" "0" "11" "9" "12" "10" "4" "0" "9"
## [71] "9" "0" "14" "11" "10" "7" "0" "17" "9" "16" "7" "7" "5" "0"
## [85] "0" "10" "10" "9" "14" "17" "11" "0" "0" "12" "6" "14" "8" "11"
## [99] "0" "0" "12" "6" "10" "14" "8" "13" "12" "8" "11" "8" "14" "11"
## [113] "8" "0" "0" "10" "13" "13" "5" "15" "0" "11" "8" "12" "10" "9"
## [127] "6" "7" "10" "6" "8" "0" "16" "0" "0" "0" "11" "12" "7" "0"
## [141] "4" "7" "7" "0" "10" "10" "12" "10" "0" "12" "12" "6" "14" "7"
## [155] "0" "6" "18" "8" "12" "15" "13" "0" "10" "11" "10" "13" "8" "0"
## [169] "6" "8" "8" "0" "9" "13" "0" "16" "10" "9" "10" "5" "11" "0"
## [183] "15" "11" "13" "14" "11" "2" "12" "11" "8" "13" "8" "9" "12" "9"
## [197] "7" "12" "9" "8" "9" "6" "11" "11" "9" "10" "12" "12" "10" "6"
## [211] "9" "14" "5" "11" "12" "8" "9" "11" "5" "13" "10" "11" "9" "9"
## [225] "10" "5" "8" "10" "7" "10" "12" "10" "7" "13" "11" "11" "10" "9"
## [239] "8" "12" "11" "7" "10" "12" "29" "5" "13" "9" "9" "12" "8"This works by going through each entry and doing the conditional on the value! Let’s now write over our old column and in the same step make everything a number.
And Alas!
## [1] 9 10 10 11 0 0 8 6 12 6 0 0 10 0 7 16 10 18 15 7 8 10 11
## [24] 8 12 6 0 17 0 0 10 15 15 8 6 13 16 0 7 7 10 0 12 5 11 9
## [47] 0 21 0 16 17 11 0 13 0 15 6 15 9 16 10 18 0 11 9 12 10 4 0
## [70] 9 9 0 14 11 10 7 0 17 9 16 7 7 5 0 0 10 10 9 14 17 11 0
## [93] 0 12 6 14 8 11 0 0 12 6 10 14 8 13 12 8 11 8 14 11 8 0 0
## [116] 10 13 13 5 15 0 11 8 12 10 9 6 7 10 6 8 0 16 0 0 0 11 12
## [139] 7 0 4 7 7 0 10 10 12 10 0 12 12 6 14 7 0 6 18 8 12 15 13
## [162] 0 10 11 10 13 8 0 6 8 8 0 9 13 0 16 10 9 10 5 11 0 15 11
## [185] 13 14 11 2 12 11 8 13 8 9 12 9 7 12 9 8 9 6 11 11 9 10 12
## [208] 12 10 6 9 14 5 11 12 8 9 11 5 13 10 11 9 9 10 5 8 10 7 10
## [231] 12 10 7 13 11 11 10 9 8 12 11 7 10 12 29 5 13 9 9 12 8Let’s now clean up the Gender column, first let’s look at it
## [1] "FEMALE" "FEMALE" "FEMALE" "MALE" "MALE" "female" "FEMALE"
## [8] "female" "MALE" "male" "FEMALE" "FEMALE" "FEMALE" "FEMALE"
## [15] "FEMALE" "FEMALE" "FEMALE" "male" "FEMALE" "FEMALE" "FEMALE"
## [22] "FEMALE" "FEMALE" "FEMALE" "MALE" "FEMALE" "MALE" "FEMALE"
## [29] "FEMALE" "" "MALE" "FEMALE" "female" "female" "FEMALE"
## [36] "female " "female" "Female" "FEMALE" "FEMALE" "MALE" "MALE"
## [43] "FEMALE" "FEMALE" "FEMALE" "MALE" "FEMALE" "FEMALE" "female"
## [50] "MALE" "MALE" "FEMALE" "FEMALE" "FEMALE" "MALE" "FEMALE"
## [57] "MALE" "FEMALE" "FEMALE" "FEMALE" "female" "FEMALE" NA
## [64] "Male" "Female" "MALE" "MALE" "FEMALE" "FEMALE" "FEMALE"
## [71] "MALE" "FEMALE" "FEMALE" "MALE" "MALE" "female" "FEMALE"
## [78] "female" "FEMALE" "female " "FEMALE" "FEMALE" "FEMALE" "FEMALE"
## [85] "FEMALE" "female" "MALE" "FEMALE" "FEMALE" "MALE" "FEMALE"
## [92] "m" "female" "male" "FEMALE" "FEMALE" "Female" "female"
## [99] "FEMALE" "FEMALE" "MALE" "female" "FEMALE" "MALE" "MALE"
## [106] "male" "FEMALE" "MALE" "female" "female" "female" "Male"
## [113] "FEMALE" "FEMALE" "FEMALE" "FEMALE" "Male" "FEMALE" "FEMALE"
## [120] "MALE" "FEMALE" "male" "FEMALE" "FEMALE" "MALE" "FEMALE"
## [127] "FEMALE" "FEMALE" "Female" "FEMALE" "FEMALE" "female" "female"
## [134] "FEMALE" "FEMALE" "FEMALE" "MALE" "MALE" "FEMALE" "FEMALE"
## [141] "MALE" "Male" "FEMALE" "female" "FEMALE" "MALE" "FEMALE"
## [148] "FEMALE" "FEMALE" "FEMALE" "MALE" "FEMALE" "male" "FEMALE"
## [155] "FEMALE" "FEMALE" "male" "FEMALE" "FEMALE" "FEMALE" "MALE"
## [162] "FEMALE" "female" "FEMALE" "FEMALE" "FEMALE" "f" "FEMALE"
## [169] "MALE" "FEMALE" "male" "Male" "FEMALE" "MALE " "female "
## [176] "female" "MALE" "FEMALE" "FEMALE" "female" "FEMALE" "COUNTRY"
## [183] "MALE" "MALE" "female" "male" "MALE" "FEMALE" "MALE"
## [190] "FEMALE" "FEMALE" "Female" "FEMALE" "FEMALE" "MALE" "MALE"
## [197] "FEMALE" "MALE" "FEMALE" "FEMALE" "MALE" "Male" "MALE"
## [204] "MALE" "MALE" "MALE" "MALE" "Male" "FEMALE" "FEMALE"
## [211] "FEMALE" "MALE" "female" "FEMALE" "MALE" "FEMALE" "MALE"
## [218] "FEMALE" "MALE" "FEMALE" "FEMALE" "FEMALE" "MALE" "FEMALE"
## [225] "MALE" "FEMALE" "FEMALE" "MALE" "MALE" "FEMALE" "FEMALE"
## [232] "FEMALE" "FEMALE" "FEMALE" "MALE" "FEMALE" "FEMALE" "FEMALE"
## [239] "FEMALE" "MALE" "FEMALE" "FEMALE" "FEMALE" "MALE" "MALE"
## [246] "FEMALE" "M" "FEMALE" "FEMALE" "FEMALE" "FEMALE"##
## COUNTRY f female Female FEMALE female m M
## 1 1 1 24 5 137 3 1 1
## male Male MALE MALE
## 9 7 59 13.1.1 Cleaning Up Gender
Pretty much two answers, how do we make them all say one thing? Let’s use the stringr package for this. Import it up top.
experiment.data$Gender <- str_to_lower(experiment.data$Gender)
experiment.data$Gender <- str_replace(experiment.data$Gender,"^.*f.*$","Female")
experiment.data$Gender <- str_replace(experiment.data$Gender,"^m.*$","Male")
experiment.data$Gender <- str_replace(experiment.data$Gender,"^country$","No Response")
experiment.data$Gender[30] <- "No Response" #Something Might Be Up w this datapoint?
experiment.data$Gender[63] <- "No Response" #Something Might Be Up w this datapoint?
experiment.data$Gender <- as.factor(experiment.data$Gender)
table(experiment.data$Gender)##
## Female Male No Response
## 170 78 3#--------------------------------------------------
# Can we do same thing for AP?
experiment.data$AbsPitch## [1] "NO" "no" "NO" "NO" "NO" "NO" "no" "NO" "NO" "no" "NO"
## [12] "NO" "NO" "NO" "NO" "NO" "NO" "no" "No" "NO" "no" "NO"
## [23] "NO" "NO" "NO" "NO" "NO" "NO" "NO" "" "No" "NO" "no"
## [34] "no" "NO" "no" "no" "no" "NO" "NO" "NO" "no" "NO" "NO"
## [45] "NO" "no" "NO" "NO" "no" "NO" "NO" "NO" "NO" "NO" "NO"
## [56] "NO" "NO" "No" "no" "NO" "No" "NO" "no" "no" "No" "NO"
## [67] "NO" "no" "no" "NO" "NO" "NO" "no" "NO" "NO" "no" "NO"
## [78] "no" "no" "NO" "NO" "NO" "NO" "NO" "NO" "no" "NO" "NO"
## [89] "NO" "NO" "NO" "NO" "no" "no" "NO" "NO" "no" "no" "NO"
## [100] "no" "NO" "no" "NO" "NO" "NO" "no" "NO" "NO" "no" "no"
## [111] "no" "no" "NO" "NO" "NO" "NO" "no" "NO" "no" "NO" "NO"
## [122] "NO" "NO" "No" "NO" "NO" "no" "NO" "no" "NO" "NO" "no"
## [133] "no" "NO" "NO" "NO" "NO" "NO" "NO" "NO" "No" "No" "NO"
## [144] "no" "NO" "NO" "NO" "NO" "NO" "no" "NO" "NO" "no" "NO"
## [155] "Non" "YES" "NO" "NO" "NO" "NO" "NO" "NO" "no" "NO" "NO"
## [166] "NO" "no" "NO" "NO" "no" "no" "NO" "NO" "no" "No" "no"
## [177] "NO" "NO" "no" "non" "NO" "NO" "NO" "NO" "No" "no" "NO"
## [188] "NO" "NO" "NO" "NO" "NO" "NO" "NO" "NO" "NO" "NO" "NO"
## [199] "no" "NO" "NO" "No" "NO" "No" "NO" "NO" "NO" "no" "NO"
## [210] "NO" "NO" "NO" "NO" "No" "No" "NO" "NO" "NO" "NO" "NO"
## [221] "NO" "NO" "NO" "NO" "NO" "NO" "NO" "NO" "NO" "NO" "NO"
## [232] "NO" "NO" "NO" "NO" "NO" "NO" "No" "No" "NO" "NO" "NO"
## [243] "no" "NO" "NO" "NO" "NO" "NO" "NO" "no" "NO"experiment.data$AbsPitch <- str_to_lower(experiment.data$AbsPitch)
experiment.data$AbsPitch <- str_replace(experiment.data$AbsPitch,"^.*n.*$","no")
experiment.data$AbsPitch[30] <- "no"
experiment.data$AbsPitch <- as.factor(experiment.data$AbsPitch)
table(experiment.data$AbsPitch)##
## no yes
## 250 13.2 Merging Data
Often we will have data from other spreadsheets we want to attach such as demographic data
to behavioral responses.
Using the data.table functionality, let’s merge our two csv files together so that we have every variable accessible to us for this analysis.
Note I like to work with the data.table package, though there are other ways to do this!
In order to do this, we need 1 shared column between the two datasets. For most psychology cases, this is probably going to be a participant ID number. Note that for this to work, you need the columns to have an exact match of name! First let’s check that they are the same!!
## [1] "Sub" "ACTIVE" "PERCEPTUAL"
## [4] "MUSICAL" "SINGING" "EMOTIONS"
## [7] "GENERAL" "inst" "Age"
## [10] "Gender" "Major" "Minor"
## [13] "SchoolUGorG" "MuscMajMinor" "BeginTrain"
## [16] "AbsPitch" "YearsPrac" "PeakPrac"
## [19] "Live12Mo" "TheoryTrain" "FormalYrs"
## [22] "NoOfInst" "ActListenMin" "SubjectNo"
## [25] "SubRaven" "RavenB1" "RavenB2"
## [28] "RavenB3" "RavenTotal" "OspanSub"
## [31] "OspanAccError" "OspanMathError" "OspanAbsoluteScore"
## [34] "OspanPartialScore" "OspanB1" "OspanB2"
## [37] "OspanB3" "OspanSpeedErrTot" "SymSub"
## [40] "SymAccErrTot" "SymSpeedErrTot" "SspanAbsScore"
## [43] "SspanPartiaScore" "SspanB1" "SspanB2"
## [46] "SspanB3" "SymmErrTot" "ToneSub"
## [49] "ToneAccErrTot" "ToneMathErrTot" "TspanAbsScore"
## [52] "TspanPartScore" "TspanB1" "TspanB2"
## [55] "TspanB3" "ToneSpeedErrTot" "BeatSub"
## [58] "BeatTotal" "BeatAcc" "MelodicSub"
## [61] "MelodicTot" "MelodicAcc" "Native"
## [64] "Paid" "jv_NoGMSI" "jv_nonNative"
## [67] "jv_noOspan"## [1] "tmp.dat.subject.1." "X1" "X2"
## [4] "X3" "X4" "X5"
## [7] "X6" "X7" "X8"
## [10] "X9" "X10" "X11"
## [13] "X12" "X13" "X14"
## [16] "X15" "X16" "X17"
## [19] "X18" "X19" "X20"
## [22] "X21" "X22" "X23"
## [25] "X24" "X25" "X26"
## [28] "X27" "X28" "X29"
## [31] "X30" "X31" "X32"
## [34] "X33" "X34" "X35"
## [37] "X36" "X37" "X38"First off our subject ID columns are not the same. Let’s swap that.
## [1] "tmp.dat.subject.1." "X1" "X2"
## [4] "X3" "X4" "X5"
## [7] "X6" "X7" "X8"
## [10] "X9" "X10" "X11"
## [13] "X12" "X13" "X14"
## [16] "X15" "X16" "X17"
## [19] "X18" "X19" "X20"
## [22] "X21" "X22" "X23"
## [25] "X24" "X25" "X26"
## [28] "X27" "X28" "X29"
## [31] "X30" "X31" "X32"
## [34] "X33" "X34" "X35"
## [37] "X36" "X37" "X38"## [1] "Sub" "ACTIVE" "PERCEPTUAL"
## [4] "MUSICAL" "SINGING" "EMOTIONS"
## [7] "GENERAL" "inst" "Age"
## [10] "Gender" "Major" "Minor"
## [13] "SchoolUGorG" "MuscMajMinor" "BeginTrain"
## [16] "AbsPitch" "YearsPrac" "PeakPrac"
## [19] "Live12Mo" "TheoryTrain" "FormalYrs"
## [22] "NoOfInst" "ActListenMin" "SubjectNo"
## [25] "SubRaven" "RavenB1" "RavenB2"
## [28] "RavenB3" "RavenTotal" "OspanSub"
## [31] "OspanAccError" "OspanMathError" "OspanAbsoluteScore"
## [34] "OspanPartialScore" "OspanB1" "OspanB2"
## [37] "OspanB3" "OspanSpeedErrTot" "SymSub"
## [40] "SymAccErrTot" "SymSpeedErrTot" "SspanAbsScore"
## [43] "SspanPartiaScore" "SspanB1" "SspanB2"
## [46] "SspanB3" "SymmErrTot" "ToneSub"
## [49] "ToneAccErrTot" "ToneMathErrTot" "TspanAbsScore"
## [52] "TspanPartScore" "TspanB1" "TspanB2"
## [55] "TspanB3" "ToneSpeedErrTot" "BeatSub"
## [58] "BeatTotal" "BeatAcc" "MelodicSub"
## [61] "MelodicTot" "MelodicAcc" "Native"
## [64] "Paid" "jv_NoGMSI" "jv_nonNative"
## [67] "jv_noOspan"setnames(item.level.data,"tmp.dat.subject.1.","SubjectNo")
setnames(experiment.data,"Sub","SubjectNo") # Make this clearer!!! If you need to do more than 1, use the c() operator! Now if we look at this column, it’s all messe up. The code below fixes it, if you want to learn more about regex, check it out if not, just skip below.
item.level.data$SubjectNo <- str_replace_all(string = item.level.data$SubjectNo, pattern = ".csv", replacement = "")
item.level.data$SubjectNo <- str_replace_all(string = item.level.data$SubjectNo, pattern = "C", replacement = "")
item.level.data$SubjectNo <- str_replace_all(string = item.level.data$SubjectNo, pattern = "M", replacement = "")
item.level.data$SubjectNo <- str_replace_all(string = item.level.data$SubjectNo, pattern = "CM", replacement = "")
item.level.data$SubjectNo <- as.numeric(item.level.data$SubjectNo)Let’s just quickly check to see if all the subject numbers make sense
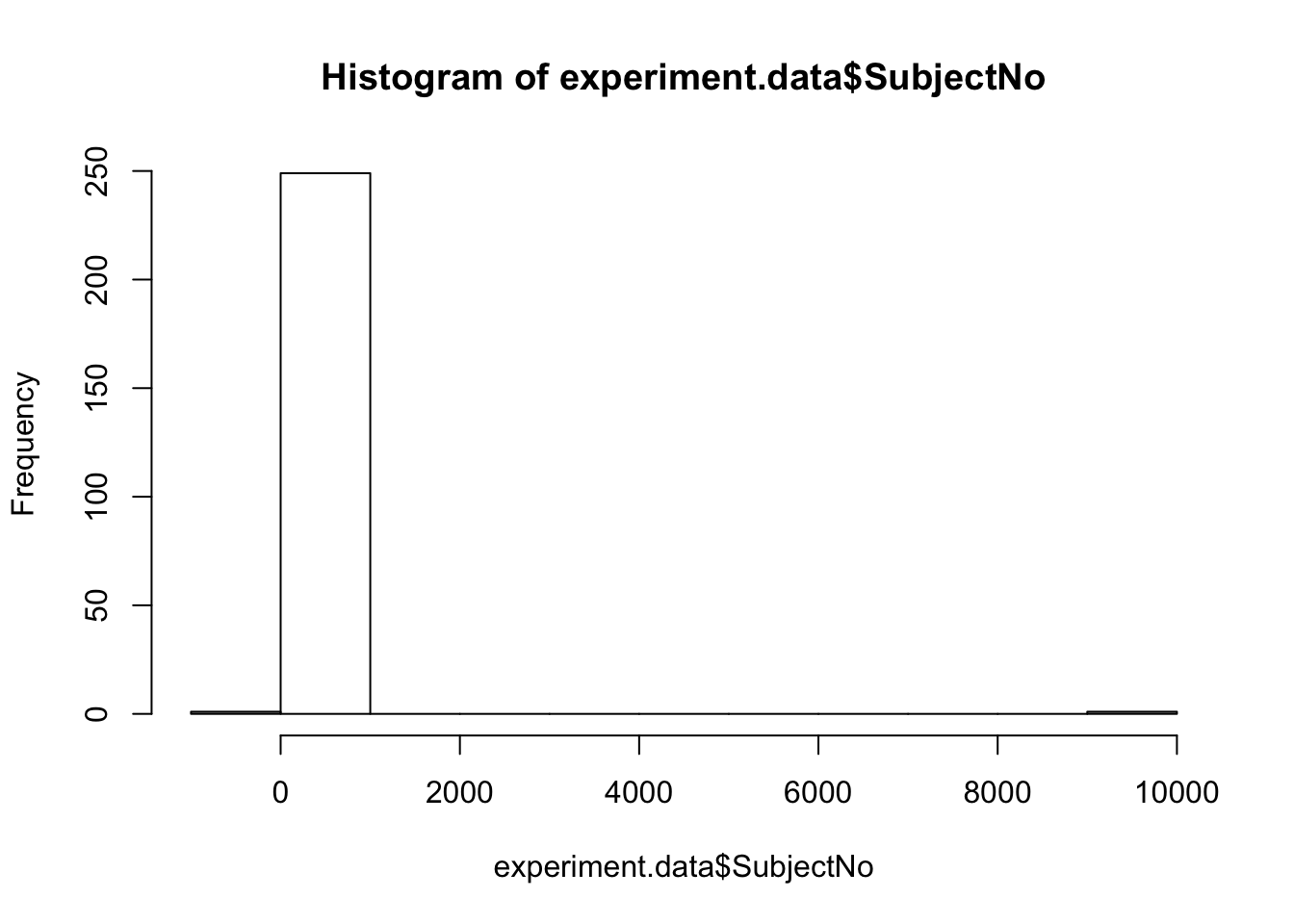
Notice that the variable of SubjectNo has negative values and placeholders!
We can use R’s indexing function to find these entries.
# Drop those
experiment.data <- experiment.data[experiment.data$SubjectNo > 0 & experiment.data$SubjectNo < 1000,]Note this works because the SubjectNo variable is numeric
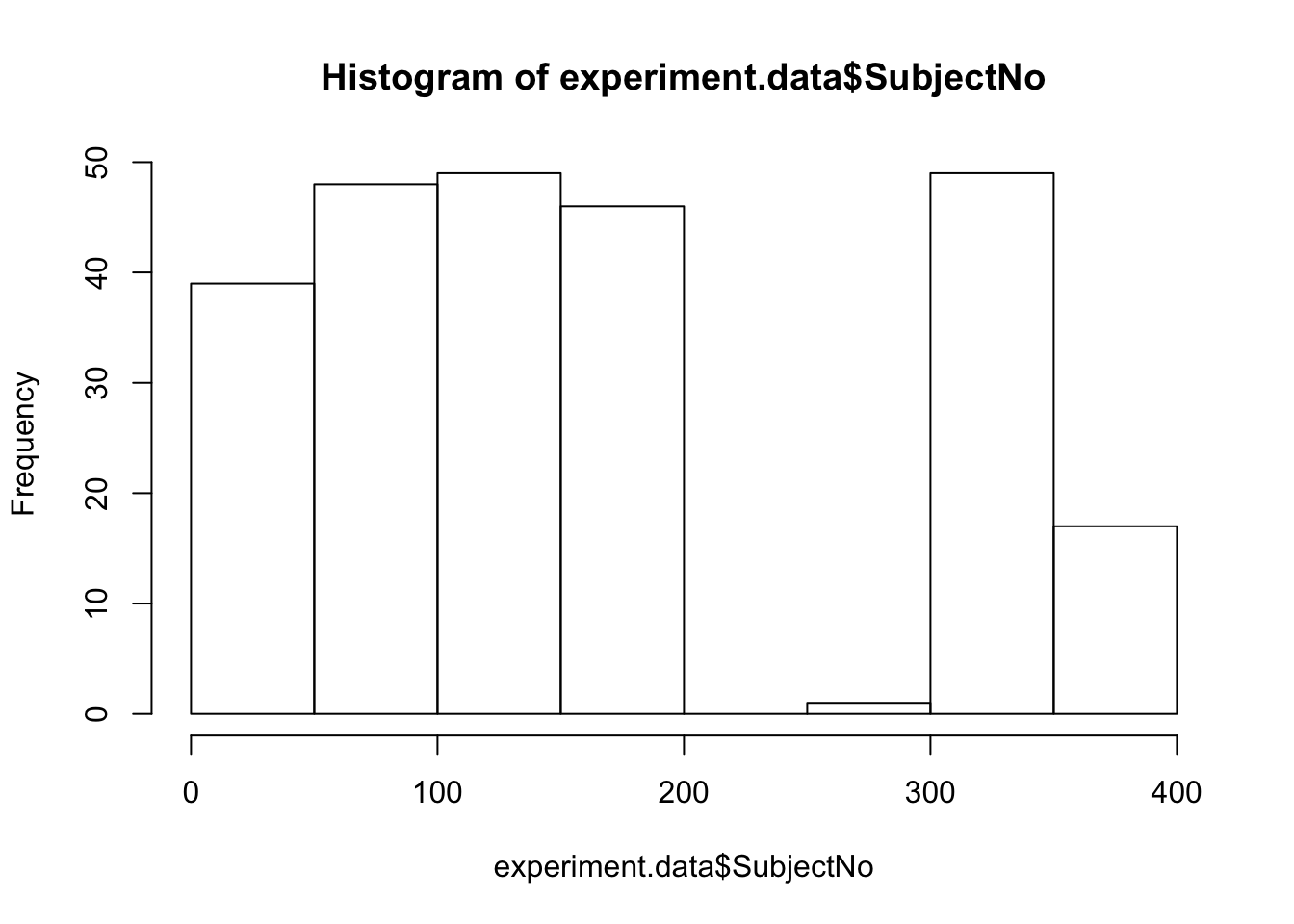
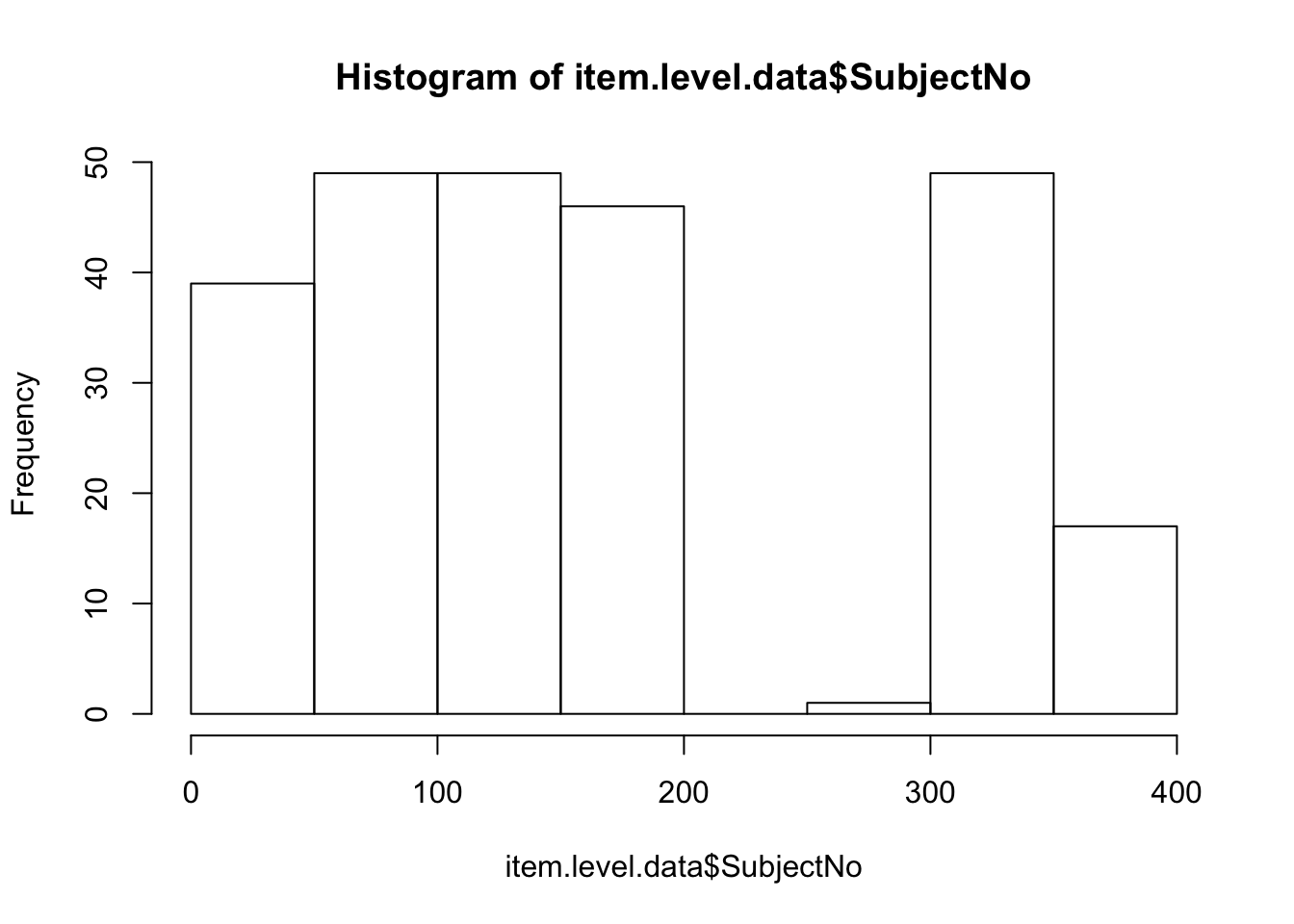
Ok, finally we merge our datasets. What we are doing here is called an “inner join” Here we will keep all of the ROWS of the dataset in the middle of the command Note we need to swap over our key to be a character value.
item.level.data <- data.table(item.level.data)
experiment.data <- data.table(experiment.data)
item.level.data## SubjectNo X1 X2 X3 X4 X5 X6 X7 X8 X9 X10 X11 X12 X13 X14 X15 X16 X17
## 1: 100 2 1 5 4 1 5 5 1 4 5 7 1 5 4 3 5 6
## 2: 101 7 5 7 7 3 7 7 4 7 2 6 4 5 5 5 4 6
## 3: 102 4 2 5 4 3 7 4 5 3 4 6 3 2 4 4 5 2
## 4: 103 5 7 7 5 5 7 7 5 7 6 7 2 5 6 5 4 4
## 5: 104 2 1 1 2 1 4 1 5 3 5 6 2 5 6 6 6 1
## ---
## 246: 95 3 2 6 5 3 2 3 3 4 2 6 6 5 6 6 6 3
## 247: 96 5 1 3 5 5 5 6 5 3 5 6 4 2 5 5 5 1
## 248: 97 5 2 4 4 2 3 5 4 2 5 6 4 3 5 5 3 6
## 249: 98 6 2 7 7 6 7 5 3 4 4 7 6 6 6 6 6 7
## 250: 99 5 4 5 5 6 7 7 4 7 2 4 5 2 5 5 4 1
## X18 X19 X20 X21 X22 X23 X24 X25 X26 X27 X28 X29 X30 X31 X32 X33 X34
## 1: 2 2 2 3 3 3 4 2 2 7 2 5 5 2 1 6 7
## 2: 6 2 1 3 5 1 1 2 3 4 2 4 3 3 2 7 7
## 3: 4 2 1 2 6 4 3 2 2 2 1 2 3 2 2 5 6
## 4: 4 2 1 1 1 3 3 1 1 2 1 5 1 5 4 7 3
## 5: 5 1 1 1 1 1 1 1 3 2 1 1 5 7 5 1 6
## ---
## 246: 6 2 2 1 1 1 1 1 6 6 3 6 6 2 2 2 6
## 247: 6 1 1 1 1 1 1 1 2 6 1 1 6 6 5 2 6
## 248: 5 6 7 3 5 1 5 2 2 5 2 2 4 5 3 5 6
## 249: 7 6 3 7 5 4 4 2 6 6 5 7 7 7 3 6 7
## 250: 5 1 1 2 3 1 1 1 6 5 2 2 3 7 6 6 7
## X35 X36 X37 X38
## 1: 7 2 4 7
## 2: 7 4 6 7
## 3: 7 5 5 7
## 4: 7 7 7 7
## 5: 6 3 5 7
## ---
## 246: 6 5 5 5
## 247: 7 2 5 6
## 248: 5 5 4 5
## 249: 7 5 6 7
## 250: 7 2 6 7## SubjectNo X1 X2 X3 X4 X5 X6 X7 X8 X9 X10 X11 X12 X13 X14 X15 X16 X17
## 1: 10 2 2 5 6 5 4 7 3 4 6 7 4 6 6 5 4 4
## 2: 11 4 2 4 5 2 5 6 5 4 6 6 3 6 5 5 5 5
## 3: 12 4 2 6 5 2 6 5 2 7 6 6 5 3 6 6 6 6
## 4: 13 5 2 3 7 2 5 5 1 3 4 6 2 4 6 6 5 3
## 5: 14 6 1 6 6 5 6 6 6 6 7 7 6 6 6 6 7 6
## ---
## 245: 363 6 1 5 2 1 7 2 2 1 7 6 3 5 6 6 6 7
## 246: 364 5 3 6 4 3 6 4 4 6 7 5 6 6 6 6 7 7
## 247: 365 3 1 5 2 1 5 5 2 1 6 6 6 5 6 5 6 5
## 248: 366 5 3 5 4 2 5 5 3 3 5 6 5 6 5 6 6 5
## 249: 367 7 5 7 7 7 6 6 7 7 7 7 6 7 6 7 7 5
## X18 X19 X20 X21 X22 X23 X24 X25 X26 X27 X28 X29 X30 X31 X32 X33 X34
## 1: 5 2 1 5 7 6 5 2 4 6 4 3 4 6 5 7 7
## 2: 6 2 2 2 5 1 3 1 4 5 2 2 2 5 2 5 5
## 3: 5 4 2 4 3 1 5 2 7 6 5 6 6 6 4 5 6
## 4: 6 3 4 6 3 1 6 3 5 5 2 2 2 2 1 6 6
## 5: 6 1 1 1 1 1 1 1 3 1 5 1 2 6 6 7 7
## ---
## 245: 6 7 2 6 5 1 6 2 6 7 7 6 5 2 1 5 6
## 246: 4 7 6 7 5 4 6 3 5 7 6 1 7 6 5 5 6
## 247: 6 6 3 6 3 1 6 2 3 4 2 4 4 2 2 6 6
## 248: 6 5 4 5 3 1 5 3 5 6 4 5 3 4 3 5 6
## 249: 6 7 7 7 7 6 6 3 4 7 6 6 2 6 3 7 7
## X35 X36 X37 X38 ACTIVE PERCEPTUAL MUSICAL SINGING EMOTIONS GENERAL
## 1: 6 6 6 7 38 47 28 33 39 75
## 2: 6 5 5 6 37 47 16 22 32 67
## 3: 7 2 4 6 39 49 21 40 30 82
## 4: 7 5 5 5 33 42 26 19 34 68
## 5: 6 6 7 2 48 57 7 24 35 60
## ---
## 245: 6 5 7 6 27 52 29 34 35 88
## 246: 6 5 6 7 41 54 38 37 35 96
## 247: 5 3 6 6 25 51 27 21 32 61
## 248: 6 5 7 5 35 50 26 30 34 82
## 249: 7 6 7 7 59 58 43 34 41 103
## inst Age Gender Major
## 1: clarinet 29 Female BUSINESS MANAGEMENT
## 2: bass 22 Female psychology
## 3: Voice 19 Female COMMUNICATION STUDIES
## 4: saxophone 23 Male Mechanical Engineering
## 5: none 23 Male Interdisciplinary Studies
## ---
## 245: voice 21 Male psychology
## 246: voice 22 Female Psychology and English Literature
## 247: flute 22 Female PSYCHOLOGY
## 248: saxophone 21 Female psychology
## 249: Bassoon 23 Female Bassoon Performance
## Minor SchoolUGorG MuscMajMinor BeginTrain
## 1: HUMAN RESOURCE MANAGEMENT 1 1 9
## 2: <NA> 1 1 10
## 3: SOCIAL WORK 1 1 10
## 4: <NA> 1 1 11
## 5: Anthropology 1 1 0
## ---
## 245: 1 1 13
## 246: Sociology 1 1 9
## 247: CHEMISTRY 1 1 9
## 248: theatre 1 1 12
## 249: 2 2 8
## AbsPitch YearsPrac PeakPrac Live12Mo TheoryTrain FormalYrs NoOfInst
## 1: no 4 5 9 4 4 1
## 2: no 1 2 15 0 1 0
## 3: no 3 1 3 0 3 1
## 4: no 7 1 2 0 7 2
## 5: no 0 0 20 0 0 0
## ---
## 245: no 9 2 3 0 9 1
## 246: no 14 2 12 2 8 2
## 247: no 8 1 3 0 9 1
## 248: no 4 1 9 0 3 2
## 249: no 12 5 16 6 9 2
## ActListenMin i.SubjectNo SubRaven RavenB1 RavenB2 RavenB3 RavenTotal
## 1: 60 10 10 10 8 10 28
## 2: 60 11 11 7 10 6 23
## 3: 180 12 12 3 7 7 17
## 4: 45 13 13 10 10 11 31
## 5: 120 14 14 7 8 5 20
## ---
## 245: 6 363 363 6 7 6 19
## 246: 160 364 364 7 8 8 23
## 247: 10 365 365 10 10 9 29
## 248: 35 366 366 10 7 9 26
## 249: 240 367 367 7 6 6 19
## OspanSub OspanAccError OspanMathError OspanAbsoluteScore
## 1: 10 8 12 29
## 2: 11 7 12 20
## 3: 12 7 7 48
## 4: 13 1 2 68
## 5: 14 2 2 33
## ---
## 245: 363 2 4 18
## 246: 364 1 1 68
## 247: 365 2 8 4
## 248: 366 4 7 38
## 249: 367 1 2 54
## OspanPartialScore OspanB1 OspanB2 OspanB3 OspanSpeedErrTot SymSub
## 1: 47 21 14 12 4 10
## 2: 41 11 18 12 5 11
## 3: 65 21 23 21 0 12
## 4: 73 25 23 25 1 13
## 5: 50 17 16 17 0 14
## ---
## 245: 28 11 8 9 2 363
## 246: 72 22 25 25 0 364
## 247: 21 10 5 6 6 365
## 248: 58 22 18 18 3 366
## 249: 66 25 22 19 1 367
## SymAccErrTot SymSpeedErrTot SspanAbsScore SspanPartiaScore SspanB1
## 1: 1 0 10 19 8
## 2: 4 1 15 29 11
## 3: 2 0 21 34 10
## 4: 1 0 27 34 13
## 5: 0 0 15 26 8
## ---
## 245: 0 0 7 21 8
## 246: 0 0 32 38 14
## 247: 0 0 18 28 9
## 248: 2 1 33 40 14
## 249: 2 0 17 28 10
## SspanB2 SspanB3 SymmErrTot ToneSub ToneAccErrTot ToneMathErrTot
## 1: 4 7 1 10 8 9
## 2: 13 5 5 11 5 5
## 3: 13 11 2 12 8 8
## 4: 10 11 1 13 1 2
## 5: 8 10 0 14 4 5
## ---
## 245: 7 6 0 363 7 8
## 246: 13 11 0 364 3 5
## 247: 8 11 0 365 3 4
## 248: 13 13 3 366 6 9
## 249: 10 8 2 367 2 2
## TspanAbsScore TspanPartScore TspanB1 TspanB2 TspanB3 ToneSpeedErrTot
## 1: 9 36 13 6 17 1
## 2: 21 47 14 19 14 0
## 3: 27 55 17 21 17 0
## 4: 37 63 20 22 21 1
## 5: 10 31 8 9 14 1
## ---
## 245: 6 37 15 13 9 1
## 246: 55 67 22 24 21 2
## 247: 21 38 9 13 16 1
## 248: 32 56 17 16 23 3
## 249: 58 67 23 23 21 0
## BeatSub BeatTotal BeatAcc MelodicSub MelodicTot MelodicAcc Native
## 1: 10 11 0.61 10 7 0.54 1
## 2: 11 10 0.56 11 7 0.54 1
## 3: 12 14 0.78 12 6 0.46 1
## 4: 13 13 0.72 13 10 0.77 1
## 5: 14 14 0.78 14 5 0.38 1
## ---
## 245: 363 16 0.89 363 7 0.54 1
## 246: 364 11 0.61 364 7 0.54 1
## 247: 365 10 0.56 365 10 0.77 1
## 248: 366 9 0.50 366 9 0.69 1
## 249: 367 12 0.67 367 9 0.69 1
## Paid jv_NoGMSI jv_nonNative jv_noOspan
## 1: 1 0 0 0
## 2: 1 0 0 0
## 3: 1 0 0 0
## 4: 1 0 0 0
## 5: 1 0 0 0
## ---
## 245: 2 0 0 0
## 246: 2 0 0 0
## 247: 2 0 0 0
## 248: 2 0 0 0
## 249: 2 0 0 0Let’s reorganize our columns so individual stuff is at the back We could do this with data.table, but it’s a different syntax so let’s swap back Normally you try to stick to minimal switching, but we’re just taking a big tour du R right now and learning to think
3.3 Checking for Univariate Outliers
For this example, let’s imagine a univariate outlier is one with a zscore greater than 3. While we could write a bit of code to look for this, let’s use the pairs.panels() function in the psych pacakge to just get used to looking at our data The function is not the biggest fan of huge datasets, so let’s index our dataset to only grab what we need. Try to change the values and look at variables of interest.
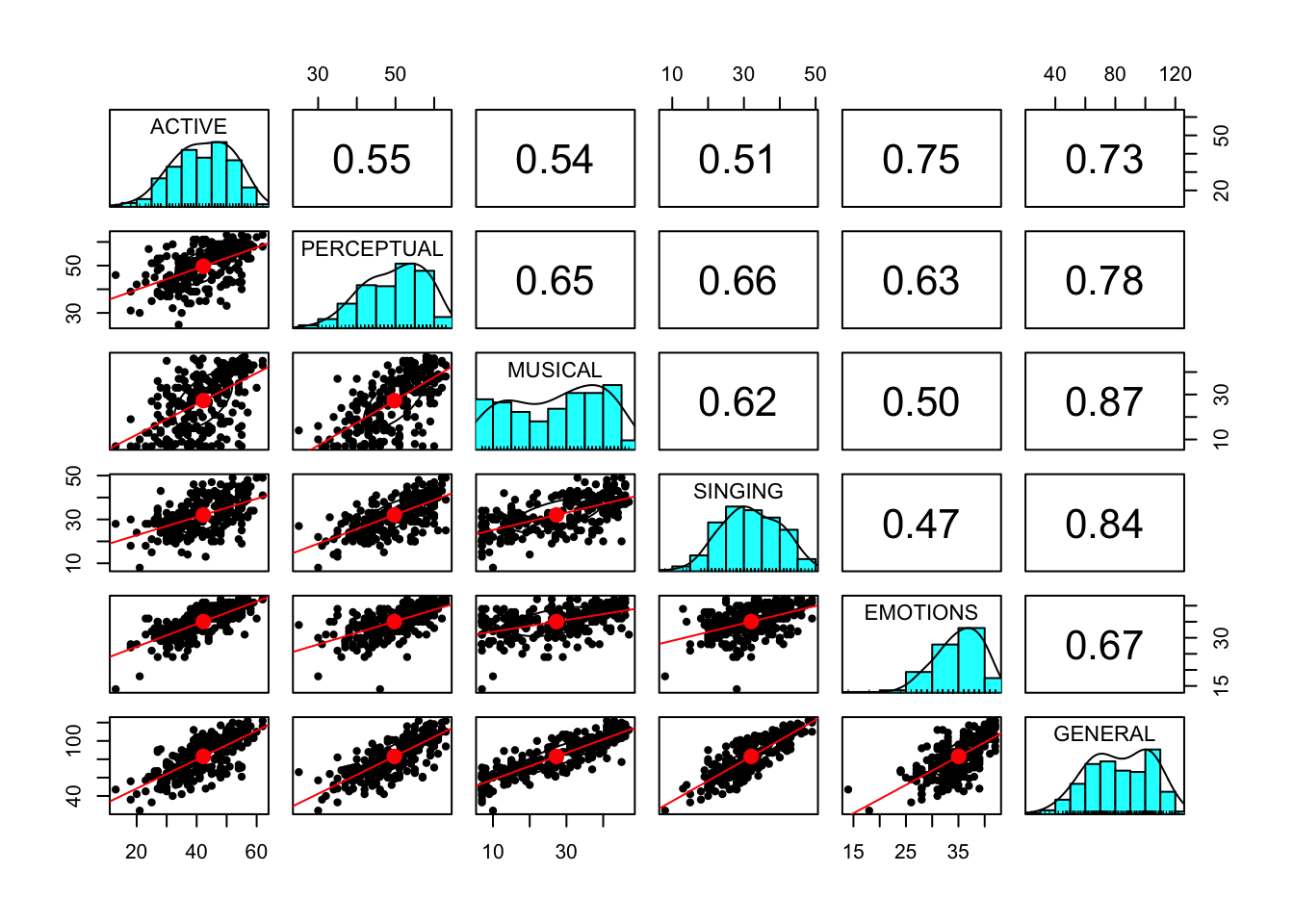
But of course we need to look at numbers in terms of their zscores! Let’s first standardize our entire dataset using the apply function Note we only can do this on numeric values!
The apply function takes 3 argument The first is what you want to manipulate, the second is if it’s rows 1 or columns 2 (remeber this because it’s always rows then columns!), and the function. You can even write your own (though we’ll get to functions later)
gmsi.z.scores <- apply(exp.data[2:7],2,scale)
exp.data.with.z <- cbind(exp.data, gmsi.z.scores)
#Now we can index this to find values above whatever theshold we want!
table(gmsi.z.scores > 2)##
## FALSE TRUE
## 1487 6gmsi.z.indexer <- gmsi.z.scores > 2
gmsi.z.scores[gmsi.z.indexer] # See what they are, find them , decide to get rid of## [1] NA 2.053441 2.053441 2.142711 2.016431 2.142711 2.1427113.3.1 Checking for Multivariate Outliers
A bit tricker, I leanred how to do this off a blog post.
gmsi.responses <- exp.data[,c(63:100)]
mahal <- mahalanobis(gmsi.responses,
colMeans(gmsi.responses, na.rm = TRUE),
cov(gmsi.responses,
use = "pairwise.complete.obs")) ## Create Distance Measures
cutoff <- qchisq(.999, ncol(gmsi.responses)) ## Create cutoff object .001 signifiance and DF = obs
summary(mahal < cutoff) ## 11 Subjects greater than 70 cutoff ## Mode FALSE TRUE
## logical 11 238# Add On Variables
exp.data$mahal <- mahal
exp.data <- data.table(exp.data) # To use easier indexer, needs data.table
exp.data[exp.data$mahal < cutoff]## SubjectNo ACTIVE PERCEPTUAL MUSICAL SINGING EMOTIONS GENERAL
## 1: 10 38 47 28 33 39 75
## 2: 11 37 47 16 22 32 67
## 3: 12 39 49 21 40 30 82
## 4: 13 33 42 26 19 34 68
## 5: 15 47 55 8 24 36 65
## ---
## 234: 363 27 52 29 34 35 88
## 235: 364 41 54 38 37 35 96
## 236: 365 25 51 27 21 32 61
## 237: 366 35 50 26 30 34 82
## 238: 367 59 58 43 34 41 103
## inst Age Gender Major
## 1: clarinet 29 Female BUSINESS MANAGEMENT
## 2: bass 22 Female psychology
## 3: Voice 19 Female COMMUNICATION STUDIES
## 4: saxophone 23 Male Mechanical Engineering
## 5: piano 19 Female marketing
## ---
## 234: voice 21 Male psychology
## 235: voice 22 Female Psychology and English Literature
## 236: flute 22 Female PSYCHOLOGY
## 237: saxophone 21 Female psychology
## 238: Bassoon 23 Female Bassoon Performance
## Minor SchoolUGorG MuscMajMinor BeginTrain
## 1: HUMAN RESOURCE MANAGEMENT 1 1 9
## 2: <NA> 1 1 10
## 3: SOCIAL WORK 1 1 10
## 4: <NA> 1 1 11
## 5: <NA> 1 1 0
## ---
## 234: 1 1 13
## 235: Sociology 1 1 9
## 236: CHEMISTRY 1 1 9
## 237: theatre 1 1 12
## 238: 2 2 8
## AbsPitch YearsPrac PeakPrac Live12Mo TheoryTrain FormalYrs NoOfInst
## 1: no 4 5 9 4 4 1
## 2: no 1 2 15 0 1 0
## 3: no 3 1 3 0 3 1
## 4: no 7 1 2 0 7 2
## 5: no 0 0 9 0 0 0
## ---
## 234: no 9 2 3 0 9 1
## 235: no 14 2 12 2 8 2
## 236: no 8 1 3 0 9 1
## 237: no 4 1 9 0 3 2
## 238: no 12 5 16 6 9 2
## ActListenMin i.SubjectNo SubRaven RavenB1 RavenB2 RavenB3 RavenTotal
## 1: 60 10 10 10 8 10 28
## 2: 60 11 11 7 10 6 23
## 3: 180 12 12 3 7 7 17
## 4: 45 13 13 10 10 11 31
## 5: 60 15 15 6 10 6 22
## ---
## 234: 6 363 363 6 7 6 19
## 235: 160 364 364 7 8 8 23
## 236: 10 365 365 10 10 9 29
## 237: 35 366 366 10 7 9 26
## 238: 240 367 367 7 6 6 19
## OspanSub OspanAccError OspanMathError OspanAbsoluteScore
## 1: 10 8 12 29
## 2: 11 7 12 20
## 3: 12 7 7 48
## 4: 13 1 2 68
## 5: 15 12 15 20
## ---
## 234: 363 2 4 18
## 235: 364 1 1 68
## 236: 365 2 8 4
## 237: 366 4 7 38
## 238: 367 1 2 54
## OspanPartialScore OspanB1 OspanB2 OspanB3 OspanSpeedErrTot SymSub
## 1: 47 21 14 12 4 10
## 2: 41 11 18 12 5 11
## 3: 65 21 23 21 0 12
## 4: 73 25 23 25 1 13
## 5: 44 14 17 13 3 15
## ---
## 234: 28 11 8 9 2 363
## 235: 72 22 25 25 0 364
## 236: 21 10 5 6 6 365
## 237: 58 22 18 18 3 366
## 238: 66 25 22 19 1 367
## SymAccErrTot SymSpeedErrTot SspanAbsScore SspanPartiaScore SspanB1
## 1: 1 0 10 19 8
## 2: 4 1 15 29 11
## 3: 2 0 21 34 10
## 4: 1 0 27 34 13
## 5: 4 1 9 25 9
## ---
## 234: 0 0 7 21 8
## 235: 0 0 32 38 14
## 236: 0 0 18 28 9
## 237: 2 1 33 40 14
## 238: 2 0 17 28 10
## SspanB2 SspanB3 SymmErrTot ToneSub ToneAccErrTot ToneMathErrTot
## 1: 4 7 1 10 8 9
## 2: 13 5 5 11 5 5
## 3: 13 11 2 12 8 8
## 4: 10 11 1 13 1 2
## 5: 10 6 5 15 7 10
## ---
## 234: 7 6 0 363 7 8
## 235: 13 11 0 364 3 5
## 236: 8 11 0 365 3 4
## 237: 13 13 3 366 6 9
## 238: 10 8 2 367 2 2
## TspanAbsScore TspanPartScore TspanB1 TspanB2 TspanB3 ToneSpeedErrTot
## 1: 9 36 13 6 17 1
## 2: 21 47 14 19 14 0
## 3: 27 55 17 21 17 0
## 4: 37 63 20 22 21 1
## 5: 4 34 9 12 13 3
## ---
## 234: 6 37 15 13 9 1
## 235: 55 67 22 24 21 2
## 236: 21 38 9 13 16 1
## 237: 32 56 17 16 23 3
## 238: 58 67 23 23 21 0
## BeatSub BeatTotal BeatAcc MelodicSub MelodicTot MelodicAcc X1 X2 X3
## 1: 10 11 0.61 10 7 0.54 2 2 5
## 2: 11 10 0.56 11 7 0.54 4 2 4
## 3: 12 14 0.78 12 6 0.46 4 2 6
## 4: 13 13 0.72 13 10 0.77 5 2 3
## 5: 15 10 0.56 15 5 0.38 7 2 5
## ---
## 234: 363 16 0.89 363 7 0.54 6 1 5
## 235: 364 11 0.61 364 7 0.54 5 3 6
## 236: 365 10 0.56 365 10 0.77 3 1 5
## 237: 366 9 0.50 366 9 0.69 5 3 5
## 238: 367 12 0.67 367 9 0.69 7 5 7
## X4 X5 X6 X7 X8 X9 X10 X11 X12 X13 X14 X15 X16 X17 X18 X19 X20 X21 X22
## 1: 6 5 4 7 3 4 6 7 4 6 6 5 4 4 5 2 1 5 7
## 2: 5 2 5 6 5 4 6 6 3 6 5 5 5 5 6 2 2 2 5
## 3: 5 2 6 5 2 7 6 6 5 3 6 6 6 6 5 4 2 4 3
## 4: 7 2 5 5 1 3 4 6 2 4 6 6 5 3 6 3 4 6 3
## 5: 7 5 7 7 3 4 7 7 3 5 6 7 7 7 6 2 1 1 1
## ---
## 234: 2 1 7 2 2 1 7 6 3 5 6 6 6 7 6 7 2 6 5
## 235: 4 3 6 4 4 6 7 5 6 6 6 6 7 7 4 7 6 7 5
## 236: 2 1 5 5 2 1 6 6 6 5 6 5 6 5 6 6 3 6 3
## 237: 4 2 5 5 3 3 5 6 5 6 5 6 6 5 6 5 4 5 3
## 238: 7 7 6 6 7 7 7 7 6 7 6 7 7 5 6 7 7 7 7
## X23 X24 X25 X26 X27 X28 X29 X30 X31 X32 X33 X34 X35 X36 X37 X38
## 1: 6 5 2 4 6 4 3 4 6 5 7 7 6 6 6 7
## 2: 1 3 1 4 5 2 2 2 5 2 5 5 6 5 5 6
## 3: 1 5 2 7 6 5 6 6 6 4 5 6 7 2 4 6
## 4: 1 6 3 5 5 2 2 2 2 1 6 6 7 5 5 5
## 5: 1 1 1 1 7 3 1 1 6 5 5 7 7 5 5 7
## ---
## 234: 1 6 2 6 7 7 6 5 2 1 5 6 6 5 7 6
## 235: 4 6 3 5 7 6 1 7 6 5 5 6 6 5 6 7
## 236: 1 6 2 3 4 2 4 4 2 2 6 6 5 3 6 6
## 237: 1 5 3 5 6 4 5 3 4 3 5 6 6 5 7 5
## 238: 6 6 3 4 7 6 6 2 6 3 7 7 7 6 7 7
## mahal
## 1: 51.21448
## 2: 25.99414
## 3: 29.78012
## 4: 49.15429
## 5: 43.12685
## ---
## 234: 58.63224
## 235: 35.67417
## 236: 34.53619
## 237: 17.72908
## 238: 20.600593.3.2 Checking for Skew and Kurtosis
## X1 X2 X3 X4 X5
## -0.751959493 0.403676499 -1.078653410 -0.813983811 0.067614732
## X6 X7 X8 X9 X10
## -1.133338356 -1.056888584 0.003114209 0.174997119 -1.251450954
## X11 X12 X13 X14 X15
## -1.209870193 -0.643154118 -1.046476716 -1.023134965 -1.512836098
## X16 X17 X18 X19 X20
## -0.918246164 -0.549121749 -0.931025766 -0.607086647 -0.009797227
## X21 X22 X23 X24 X25
## -0.324554196 -0.342355019 0.502388714 -0.404073175 1.103530636
## X26 X27 X28 X29 X30
## -0.419299836 -1.204955610 -0.411695100 -0.409111747 -0.082358311
## X31 X32 X33 X34 X35
## -1.037773224 0.068284056 -1.083784471 -1.825310783 -1.693280548
## X36 X37 X38
## -0.651012271 -1.218176414 -2.364635137## X1 X2 X3 X4 X5 X6
## -0.27214952 -0.83327603 1.14629947 0.02931285 -1.30675305 0.83305054
## X7 X8 X9 X10 X11 X12
## 1.36849018 -0.92937707 -0.69656989 1.58298149 2.00805464 -0.58320443
## X13 X14 X15 X16 X17 X18
## 0.49964877 1.30350514 2.47139233 0.56622834 -0.89787441 0.80425156
## X19 X20 X21 X22 X23 X24
## -1.18236897 -1.67596746 -1.49414255 -1.24558633 -1.41680710 -1.30953622
## X25 X26 X27 X28 X29 X30
## 0.42141756 -0.69749482 0.73800105 -1.08494701 -0.90943305 -1.31818613
## X31 X32 X33 X34 X35 X36
## 0.86327735 -1.00536155 0.46640336 4.08373503 4.90078751 -0.09764595
## X37 X38
## 2.04104393 9.603892993.3.3 Exporting Data
It’s best practice to separate your cleaning and your analysis into separate scripts. Export the dataset you have into a new csv file into a directory that would make sense to someone who has never seen your project before.